Abstract
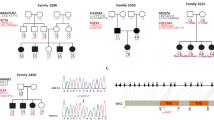
Hearing impairment is the most commonly occurring condition that affects the ability of humans to communicate 1 . More than 50% of the cases of profound early-onset deafness are caused by genetic factors 2, 3 . Over 40 loci for non-syndromic deafness have been genetically mapped, and mutations in several genes have been shown to cause hearing loss 4 . Mutations in the gene encoding connexin 26 ( GJB2 ) cause both autosomal recessive and dominant forms of hearing impairment 5, 6 . To study the possible involvement of other members of the connexin family in hereditary hearing impairment, we cloned the gene ( GJB3 ) encoding human gap junction protein β-3 using homologous EST searching and nested PCR. GJB3 was mapped to human chromosome 1p33-p35. Mutation analysis revealed that a missense mutation and a nonsense mutation of GJB3 were associated with high-frequency hearing loss in two families. Moreover, expression of Gjb3 was identified in rat inner ear tissue by RT-PCR. These findings suggest that mutations in GJB3 may be responsible for bilateral high-frequency hearing impairment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Morton, N.E. Genetic epidemiology of hearing impairment. Ann. N.Y. Acad. Sci . 630, 16–31 ( 1991).
Marazita, M.L. et al. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am. J. Med. Genet. 46, 486–491 (1993).
Reardon, W. Genetic deafness. J. Med. Genet. 29, 521 –526 (1992).
Van Camp, G., Willems, P.J. & Smith, R.J.H. Non-syndromic hearing impairment: unparalleled heterogeneity. Am. J. Hum. Genet. 60, 758– 764 (1997).
Denoyelle, F. et al. Connexin 26 gene linked to a dominant deafness. Nature 393, 319–320 ( 1998).
Kelsell, D.P. et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387, 80– 83 (1997).
Bergoffen, J. et al. Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science 262, 2039–2042 (1993).
Coucke, P. et al. Linkage of autosomal dominant hearing loss to the short arm of chromosome 1 in two families. N. Engl. J. Med. 331, 425–431 (1994).
Van Camp, G. et al. Linkage analysis of progressive hearing loss in five extended families maps the DFNA2 gene to a 1.25-Mb region on chromosome 1p. Genomics 41, 70–74 ( 1997).
van der Schroeff, J.G., van Leeuwen-Cornelisse, I., van Haeringen, A. & Went, L.N. Further evidence for localization of the gene of erythrokeratodermia variabilis. Hum. Genet. 80, 97–98 (1998).
Ben Othmane, K. et al. Localization of a gene (CMT2A) for autosomal dominant Charcot-Marie-Tooth disease type 2 to chromosome 1p and evidence of genetic heterogeneity. Genomics 17, 370– 375 (1993).
Engle, E.C., Castro, A.E., Macy, M.E., Knoll, J.H. & Beggs, A.H. A gene for isolated congenital ptosis maps to a 3-cM region within 1p32-p34.1. Am. J. Hum. Genet. 60, 1150–1157 (1997).
Kelley, P.M. et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am. J. Hum. Genet. 62, 792–799 ( 1998).
Fain, P.R., Barker, D.F. & Chance, P.F. Refined genetics mapping of X-linked Charcot-Marie-Tooth neuropathy. Am. J. Hum. Genet. 54, 229– 235 (1994).
Scott, D.A., Kraft, M.L., Stone, E.M., Sheffield, V.C. & Smith, R.J. Connexin mutations and hearing loss. Nature 391, 32 (1998).
White, T.W., Deans, M.R., Kelsell, D.P. & Paul, D.L. Connexin mutation in deafness. Nature 394, 630–631 (1988).
Ayimu, D. & Qiu, C. Pure tone threshold in 5000 normal ears. J. Audiol. Speech Pathol. 5, 144– 145 (1997).
Hull, R.H. Hearing evaluation of the elderly. in Handbook of Clinical Audiology (ed. Katz, J. ) 426–441 (Williams & Wilkins, Baltimore, 1978).
Elgoyhen, A.B., Johnson, D.S., Boulter, J., Vetter, D.E. & Heinemann, S. α9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell 79, 705–715 (1994).
Acknowledgements
This study was supported by a Chinese 863 projects grant (z19-02-02-02), the Chinese Natural Science Foundation (grant 39392902) and SmithKline Beecham-Hunan Medical University Collaborative Project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xia., Jh., Liu, Cy., Tang, Bs. et al. Mutations in the gene encoding gap junction protein β-3 associated with autosomal dominant hearing impairment. Nat Genet 20, 370–373 (1998). https://doi.org/10.1038/3845
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/3845
This article is cited by
-
Gene therapy development in hearing research in China
Gene Therapy (2020)
-
Whole-exome sequencing identifies two novel mutations in KCNQ4 in individuals with nonsyndromic hearing loss
Scientific Reports (2018)
-
ELMOD3, a novel causative gene, associated with human autosomal dominant nonsyndromic and progressive hearing loss
Human Genetics (2018)
-
Utilization of amplicon-based targeted sequencing panel for the massively parallel sequencing of sporadic hearing impairment patients from Saudi Arabia
BMC Medical Genetics (2016)
-
Distinct Expression Patterns Of Causative Genes Responsible For Hereditary Progressive Hearing Loss In Non-Human Primate Cochlea
Scientific Reports (2016)