Article Text
Abstract
FMR1 mRNA levels were determined in peripheral blood leucocytes for 48 fragile X males with methylated, full mutation alleles that are resistant to cleavage by methylation sensitive enzymes. Using quantitative (fluorescence) RT-PCR, we observed that more than half of these males produceFMR1 mRNA, with some mRNA levels approaching those found in normal subjects. In none of the samples analysed was there any evidence of premutation alleles. These results suggest that the assumed relationship between enzyme resistance andFMR1 gene silencing may not be generally valid. Despite the presence of FMR1 mRNA in some subjects, no FMRP production was detected by either immunocytochemistry or western blotting. The low/absent FMRP levels are probably a reflection of a post-trancriptional effect such as a defect in translation.
- trinucleotide repeat
- gene silencing
- quantitative RT-PCR
- neurodevelopment
Statistics from Altmetric.com
Fragile X syndrome nearly always arises as a consequence of a large expansion of a CGG trinucleotide repeat in the CG rich promoter region of the fragile X mental retardation 1 (FMR1) gene. Expansion of the CGG repeat into the full mutation range (>200 repeats) usually leads to hypermethylation of the CG rich region and transcriptional silencing.1 2 The consequent absence ofFMR1 protein (FMRP) results in fragile X syndrome.
From the general relationship between hypermethylation and silencing, hypermethylation per se (or the absence of FMRP) is often taken as evidence that the FMR1 gene is transcriptionally silent. Thus, expression of FMRP in patients with hypermethylated, full mutation alleles is usually interpreted as resulting from either a small percentage of premutation alleles within the cell population being examined or to epigenetic (methylation) mosaicism. In this regard, hypermethylation is generally defined operationally in terms of resistance to cleavage of the CG rich promoter region by methylation sensitive restriction enzymes (for example, NruI,EagI, BssHII).
In this report, we present evidence that a substantial fraction of males with full mutation alleles that are resistant to cleavage by methylation sensitive enzymes, and which produce little or no FMRP, nevertheless produce FMR1 mRNA, with some mRNA levels approaching those found in normal subjects. We show further that the mRNA is unlikely to be produced from low levels of premutation alleles. Thus, it is not generally valid to assume that enzyme resistance and/or absence of FMRP are indicative of a transcriptionally silent FMR1 gene.
Methods
Genomic DNA was isolated from peripheral blood leucocytes using Puregene kits (Gentra Inc). Allele sizes and methylation status were determined by standard Southern blot analysis, usingNruI as the methyl sensitive restriction enzyme3 and the FMR1 specific probe StB12.3.1 For the detection of premutation alleles, CGG repeat lengths were determined by PCR analysis using primers 1 and 3.4
Total RNA was isolated from peripheral blood using Purescript kits (Gentra Inc). cDNA synthesis was as described in Tassoneet al.5 Quantitative RT-PCR measurements of relative FMR1 mRNA levels were carried out using the 5′ fluorogenic RT-PCR assay.6 7 Details and probe specific sequences for theFMR1 gene and for the reference gene (glucoronidase) were as described in Tassone et al.5
FMRP expression was determined as the percentage of FMRP positive lymphocytes from blood smears using the immunocytochemical approach8 9 and anti-FMRP monoclonal antibody from hybridoma clone 1C3-a.10 Details of the method are described elsewhere.5 8 9 11 Western blot analysis on total protein isolated from lymphoblastoid cell lines was performed as described in Tassone et al.12
Results
To address the linkage between enzyme resistance and silencing, we have examined FMR1 mRNA and FMRP levels in peripheral blood leucocytes from 50 males. Two males possessed deletions that were transcriptionally silent. Forty eight subjects possessed full mutation alleles, ranging from 230 to >700 CGG repeats, which were resistant to cleavage by at least one methyl sensitive restriction enzyme, and which did not include premutation alleles (<1%) as determined by combined Southern and PCR analysis.
Results of quantitative RT-PCR measurements of relativeFMR1 mRNA levels are shown in fig 1. Surprisingly, we found that leucocytes from the majority (60%) of subjects continue to express some FMR1 mRNA, with six males (12%) having FMR1 mRNA levels in the same range as observed in normal subjects despite essentially complete resistance to cleavage by at least one restriction enzyme. Two additional males have FMR1 mRNA levels between 40-70% of normal, eight have levels between 10-40%, and 13 possess detectable levels (∼1-10%). Nineteen males (40%) with alleles of comparable size did not yield any detectableFMR1 mRNA after 50 cycles of PCR (<0.1%). Thus, more than half of the full mutation males in the current group continue to produce some FMR1 mRNA in leucocytes despite satisfying the enzymatic criteria for hypermethylated, full mutation alleles.
FMR1 mRNA levels in peripheral blood leucocytes for males with methylated, full mutation alleles, plotted as a function of the CGG repeat number (smallest allele when multiple bands are present). FMR1 mRNA levels are normalised to the mean level for normal controls.5 Grey triangles indicate positions of smallest CGG repeats for subjects with no detectable FMR1 mRNA after 50 cycles of fluorescent RT-PCR (<0.1%). Open/closed squares indicate repeat mRNA analyses from separate blood samples for two subjects.
The methylation status of additional restriction sites (EagI and BssHII) was also examined for four subjects with the highest levels ofFMR1 mRNA (fig 2). Densitometric analysis of the film presented in fig 2 generally showed the absence of unmethylated alleles (<1% of total density between 2.8 kb and 5.2 kb bands) with at least one enzyme. We did note the presence of some density (∼15%) in the BssHII lane of sample 3, with faint densities in the EagI lane of sample 3, and in the NruI andEagI lanes of sample 2. This last observation could reflect either incomplete methylation at the corresponding cleavage sites (although no unmethylated alleles were detected in the PCR assay, see below) or background contamination in those lanes. Consistent with this observation, Stögeret al 13 observed incomplete methylation at the BssHII site for alleles that were completely methylated at the NruI site (intra-allelic mosaicism of the methylation epigenotype) in addition to their observations of all or none methylation (interallelic mosaicism).
Southern blot analysis of the methylation status of the FMR1 CG rich region for four males with full mutation alleles. For each subject, 5 μg of genomic DNA isolated from peripheral blood leucocytes was digested with EcoRI and either NruI (N), EagI (E), or BssHII (B). The blot was probed with StB12.3 as previously described.3 A female (premutation) control (C) is also presented (cleavage with NruI). The positions of the normal unmethylated (2.8 kb) and normal methylated (5.2 kb) bands are indicated.
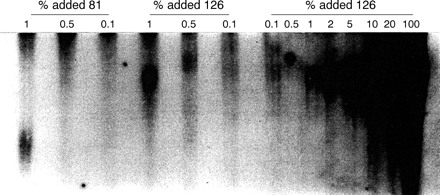
Genomic DNA from all 50 subjects (including the two deletions) was also analysed by PCR as described in Brown et al.4 None of the samples yielded any evidence of premutation alleles after 30 cycles, and for the four subjects presented in fig 2, no bands were detected after 35 cycles of PCR. To check the sensitivity of our PCR assay for low levels of premutation alleles, we performed replicate, standard PCR reactions in which a full mutation DNA sample (263 CGG repeats) was mixed with varying percentages (0.1 to 20%) of premutation alleles having either 81 or 126 repeats (fig 3). The full mutation allele was chosen based on its relatively high FMR1 mRNA level (1.28 ± 0.18, fig 2). In all PCR reactions, the total initial DNA concentration was held constant at 200 ng. In all experiments, premutation alleles were detected at the 1% level. This level of sensitivity (∼1%) using PCR has also been reported by de Graaff et al.14 Thus, our PCR protocol allows us to exclude the presence of premutation alleles at the 1% level in the full mutation samples presented in fig 1. For an additional set of controls, mRNA levels were analysed from repeat blood samples for two subjects with high expression levels (fig 1) and for three subjects with no detectable mRNA, the latter as separate PCR reactions from single mRNA isolates (data not shown). In all instances, mRNA results were highly concordant.
PCR analysis of a full mutation allele (263 CGG repeats) to which defined amounts of premutation alleles (81 or 126 CGG repeats) have been added. Percentages and allele sizes are indicated over the lanes. Repeat analyses (data not shown) always showed detection at the 1% level for both premutation alleles. Lower mixing concentrations (⩽1%) were repeated in the six lanes to the left, with lane separation to minimise lane overlap (20-24 hour exposure).
Discussion
In our previous studies of FMR1 mRNA levels in peripheral blood leucocytes with unmethylated premutation alleles, we showed that FMR1 message levels are five to ten-fold higher than normal in the upper end of the premutation range,5 despite moderate reductions in FMRP levels (by immunocytochemistry). More recently, we have extended these observations into the full mutation range for unmethylated and partially methylated alleles.15 Again,FMR1 mRNA levels were found to be increased by as much as six-fold over normal, with FMRP levels that were invariably reduced, as determined by both immunocytochemistry and western blotting, the latter using corresponding transformed lines. In this study, we have extended these observations into the full mutation range. More than half of the subjects with hypermethylated full mutation are producing significant amounts ofFMR1 mRNA, so theirFMR1 genes are clearly not silenced.
The highest FMR1 mRNA levels found to date, for either premutation or for unmethylated alleles in the full mutation range, are no more than roughly ten-fold increased over normal levels.5 15 Therefore, premutation alleles, if present at or below the 1% level, could not account forFMR1 mRNA levels that are above 10% of normal levels, roughly one third of the males in the current study. For males with relative mRNA levels in the 1-10% range (fig 1), we cannot rule out the possibility that an undetectably small fraction of unmethylated alleles (<1%) gives rise to part or all of the residual mRNA. These results, which agree with previous findings that fully methylated, full mutation alleles are mitotically stable,16 17 establish that the observed mRNA levels do not result from an undetectably small fraction of cells with premutation alleles. In addition, in a previous study, de Graaffet al 14 noted that in a lung tumour sample from a fragile X patient with a fully methylated premutation allele (∼160 repeats), 30-40% of cells were expressing FMRP by immunocytochemistry. These authors argued that such a high percentage of FMRP positive cells could not be the result of smaller, unmethylated alleles, since those alleles would not have gone undetected in their DNA assays. Although the observations of de Graaffet al 14 were based on expression patterns in tumour tissue, which may be substantially dysregulated, their results, along with those of the current work, raise an important caveat regarding the assumed relationship between resistance to enzyme cleavage and transcriptional silencing.
Thus, for many full mutation alleles that are resistant to cleavage with methylation sensitive restriction enzymes, theFMR1 gene nevertheless remains active. In fact, for six subjects, FMR1 mRNA levels are approximately equal to the mean level for normal subjects.5 However, it should be noted that in the context of full mutation alleles, “normal” levels probably represent a significant transcriptional deficit, since leucocytes with unmethylated alleles of the same length have mRNA levels that are four to six-fold increased.15
We do not yet know why some subjects in the current group produceFMR1 mRNA while others do not, although we favour the proposal that methylation of a specific subset of CpG elements is critical for silencing.13 14 18 Given the broad range of mRNA levels in fig 1 (for example, greater than 100-fold range of mRNA levels among males with methylated alleles of ∼330 repeats), it is likely that expression levels are influenced by methylation at several CpG sites. We wish to stress that we cannot distinguish between intra-allelic and interallelic mosaicism for many of the subjects represented in fig 1, particularly for those with relative mRNA levels that are below 10-20% of normal. However, for the alleles with the highest expression levels, interallelic mosaicism is unlikely, since we would have been able to detect 10-20% of completely unmethylated alleles.
In the current study, none of the subjects examined (including two in the highest mRNA group) expressed more than 10% FMRP positive lymphocytes (range 0-10%, mean 4.6%, 99% CI 2.7-6.5%), measured by immunocytochemical staining.16 17 The small percentages of FMRP(+) cells may represent, in part, intrinsic errors in the staining method, since western analysis of protein levels for lymphoblastoid lines derived from six members of the current group, two with mRNA levels in the 6-8% range (263, 363 repeats, fig 1) and four with no detectable mRNA, showed no FMRP expression.
In summary, full mutation alleles that appear to be predominantly methylated may not be silent despite low/absent FMRP expression. Thus, it is not always correct to conclude that theFMR1 gene is “silent” on the basis of either enzymatic criteria or low/absent protein levels. The low protein levels probably reflect a post-transcriptional effect such as a defect in translation.5 19 Any significant deficit in translation of the FMR1 mRNA is of central importance for therapeutic approaches aimed at recovering function from the endogenous gene, since it is not sufficient simply to reactivate the gene itself without also addressing the subsequent problem of translation. Clearly, there may be differences between leucocytes and neurones in the level of expression of theFMR1 gene, with the possibility that hypermethylated alleles are not transcribed in neuronal cells; a better understanding of this issue must await more detailed analysis of neural tissues. Furthermore, because methyl sensitive restriction enzymes test for methylation at only a few CpG positions, it is possible that silencing of the FMR1 gene is the result of focal CpG methylation events at other positions that are not probed by the enzymes. Finally, it is possible that the range of mRNA levels reflects differences in the degree of histone acetylation for equivalent epigenotypes. These possibilities are currently being investigated.
Acknowledgments
This work was supported by the Cooper/Kraff/Fishman Family Fund and Boory Family Fund (to PJH and RJH), a FRAXA Research Foundation Fellowship (to FT), National Institutes of Health Grants GM35305 (to PJH) and HD36071 (to RJH), and a Maternal Child Health Bureau grant MCJ-089413 (to (RJH).